Single compound comparisons
Contents
3.4. Single compound comparisons#
In this notebook, we compare up to 3 MD simulations of a single compound and optionally also a cheminformatics conformer generator.
This notebook refers to compound 22.
3.4.1. Imports & file inputs#
import matplotlib
%matplotlib inline
# matplotlib.use("Agg")
import matplotlib.pyplot as plt
import matplotlib.tri as tri
# set matplotlib font sizes
SMALL_SIZE = 13
MEDIUM_SIZE = 14
BIGGER_SIZE = 15
plt.rc("font", size=MEDIUM_SIZE) # controls default text sizes
plt.rc("axes", titlesize=BIGGER_SIZE) # fontsize of the axes title
plt.rc("axes", labelsize=MEDIUM_SIZE) # fontsize of the x and y labels
plt.rc("xtick", labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc("ytick", labelsize=MEDIUM_SIZE) # fontsize of the tick labels
plt.rc("legend", fontsize=MEDIUM_SIZE) # legend fontsize
plt.rc("figure", titlesize=BIGGER_SIZE) # fontsize of the figure title
DPI = 600
import mdtraj as md
import numpy as np
import scipy.cluster.hierarchy
from scipy.spatial.distance import squareform
import pandas as pd
sys.path.append(os.getcwd())
from src.pyreweight import reweight
from src.noe import compute_NOE_mdtraj, plot_NOE
from src.utils import json_load, dotdict
import src.utils
import src.dihedrals
import src.pyreweight
from sklearn.manifold import TSNE
from sklearn.cluster import DBSCAN
from sklearn.neighbors import NearestNeighbors
from sklearn.decomposition import PCA
# read in stride from config file
stride = int(snakemake.config["stride"])
stride = 1
compound_index = int(snakemake.wildcards.compound_dir)
display(Markdown(f"Analysing Compound {compound_index}"))
compound = json_load(snakemake.input.parm)
multi = compound.multi
if multi is not None:
display(Markdown("Multiple compounds detected"))
display(Markdown(print(multi)))
multi = {v: k for k, v in multi.items()}
multiple = True
else:
multiple = False
display(Markdown("Single compound only (no exp. cis/trans data known)"))
Analysing Compound 22
Single compound only (no exp. cis/trans data known)
# File inputs
methods = {}
repeats = {}
simtime = {}
solvent = {}
boosting = {}
dihe_all = {}
multi_details = {}
for i in range(3):
# Load MD details
methods[i] = snakemake.params[f"sample_{i}"]["method"]
simtime[i] = snakemake.params[f"sample_{i}"]["simtime"]
repeats[i] = snakemake.params[f"sample_{i}"]["repeats"]
solvent[i] = snakemake.params[f"sample_{i}"]["solvent"]
dihe_all[i] = src.utils.pickle_load(snakemake.input[f"red_dihe_{i}"])
if snakemake.params[f"sample_{i}"]["igamd"] != "nan":
boost = snakemake.params[f"sample_{i}"]["igamd"]
if boost == str(3):
boosting[i] = "boost: dual"
elif boost == str(2):
boosting[i] = "boost: dihe"
elif boost == str(1):
boosting[i] = "boost: totE"
else:
boosting[i] = ""
if multiple:
multi_details[i] = src.utils.pickle_load(
snakemake.input[f"multiple_{i}"]
)
multi_plots = 2
else:
multi_plots = 1
if snakemake.params[f"sample_{1}"] == snakemake.params[f"sample_{2}"]:
no_md = 2
else:
no_md = 3
if multiple:
cis = []
trans = []
for i in range(3):
cis_temp, trans_temp = src.utils.pickle_load(
snakemake.input[f"multiple_{i}"]
)
cis.append(cis_temp)
trans.append(trans_temp)
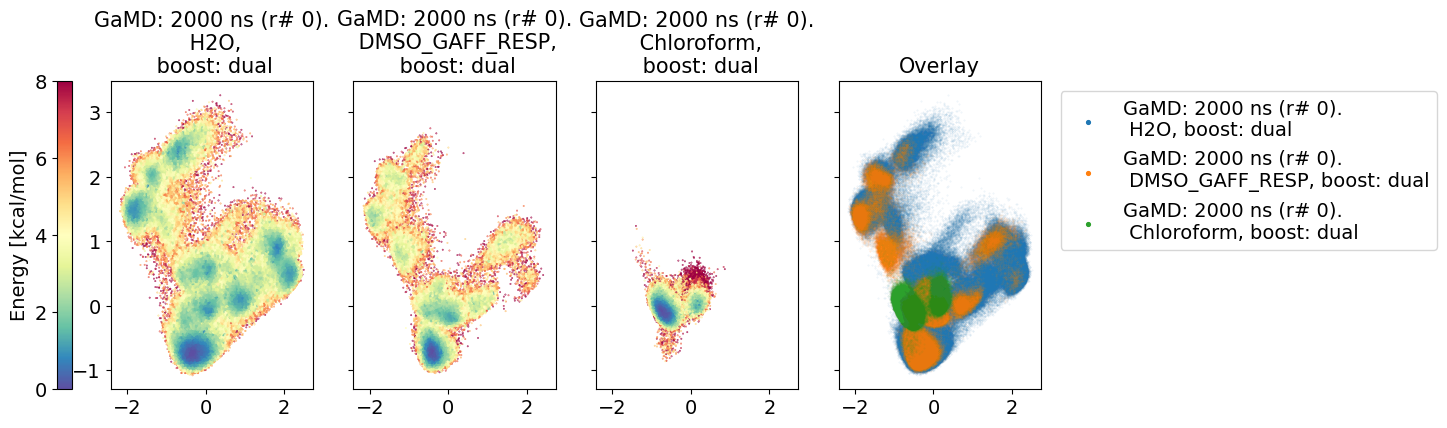
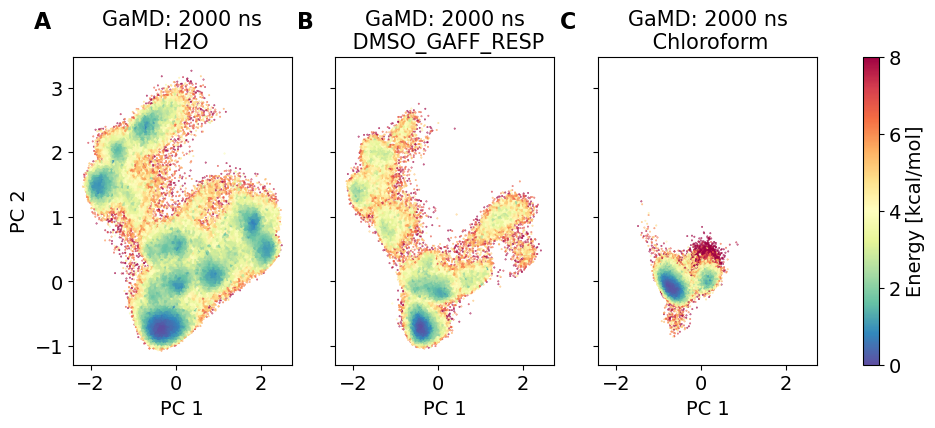
3.4.2. Dihedral PCA comparison#
The following dihedral PCA plots compare the different MD simulations. The middle and left dPCA plots are in the PCA space of the left most plot.
dihe_r = {} # This stores the reduced dihedrals
# Load pca object of reference MD simulation (0)
pca = src.utils.pickle_load(snakemake.input.dPCA_0) # PCA(n_components=2)
dihe_r[0] = pca.transform(dihe_all[0])
dihe_r[0].shape
# Now apply the same transformation to second and third
dihe_r[1] = pca.transform(dihe_all[1])
dihe_r[2] = pca.transform(dihe_all[2])
display(
Markdown(
f"The two components explain {sum(pca.explained_variance_ratio_):.2%} of the variance"
)
)
The two components explain 34.60% of the variance
# Reweighting:
weights = {}
weight_data = {}
for i in range(3):
# The following would read pre-computed weights. But does not work here b/c computed for wrong PCA space!
# weights[i] = src.utils.pickle_load(snakemake.input[f"dPCA_weights_MC_{i}"])
if methods[i] != "cMD":
weight_data[i] = np.loadtxt(snakemake.input[f"weights_{i}"])
weight_data[i] = weight_data[i][::stride]
weights[i] = reweight(
dihe_r[i],
snakemake.input[f"weights_{i}"],
"amdweight_MC",
weight_data[i],
)
else:
weights[i] = reweight(dihe_r[i], None, "noweight")
zs = np.concatenate([weights[0], weights[1], weights[2]], axis=0)
min_, max_ = zs.min(), zs.max()
min_ = 0
max_ = 8
# Plot re-weighted PCA plots with extended labels
fig, axs = plt.subplots(
1, no_md + 1, sharex="all", sharey="all", figsize=(16, 4)
)
scat = {}
for i in range(no_md):
scat[i] = axs[i].scatter(
dihe_r[i][:, 0],
dihe_r[i][:, 1],
c=weights[i],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs[i].set_title(
f"{methods[i]}: {simtime[i]} ns (r# {repeats[i]}).\n {solvent[i]},\n {boosting[i]}"
)
axs[no_md].scatter(
dihe_r[i][:, 0],
dihe_r[i][:, 1],
marker=".",
s=0.5,
alpha=0.1,
label=f"{methods[i]}: {simtime[i]} ns (r# {repeats[i]}).\n {solvent[i]}, {boosting[i]}",
rasterized=True,
)
lgnd = axs[no_md].legend(bbox_to_anchor=(1.05, 1), loc="upper left")
for handle in lgnd.legendHandles:
handle.set_sizes([30.0])
handle.set_alpha(1)
axs[no_md].set_title("Overlay")
colorbar = fig.colorbar(
scat[0],
ax=axs,
label="Energy [kcal/mol]",
location="left",
anchor=(1.5, 0),
)
fig.savefig(snakemake.output.pca_dihe, bbox_inches="tight", dpi=600)
# Publication/report quality figure:
# Plot re-weighted PCA plots
fig, axs = plt.subplots(
multi_plots,
no_md,
sharex="all",
sharey="all",
figsize=(12, 4 * multi_plots),
)
scat = {}
panel_labels = ["A", "B", "C"]
panel_labels = panel_labels[0:no_md]
for i, panel in enumerate(panel_labels):
if multiple:
# cis
scat[i] = axs.flatten()[i].scatter(
dihe_r[i][multi_details[i][0], 0],
dihe_r[i][multi_details[i][0], 1],
c=weights[i][multi_details[i][0]],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs.flatten()[i].set_title(
f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}, cis"
)
axs.flatten()[i].text(
-0.1,
1.15,
panel,
transform=axs.flatten()[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
# trans
scat[i + 2] = axs.flatten()[i + 2].scatter(
dihe_r[i][multi_details[i][1], 0],
dihe_r[i][multi_details[i][1], 1],
c=weights[i][multi_details[i][1]],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs.flatten()[i + 2].set_title(
f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}, trans"
)
axs.flatten()[i + 2].set_xlabel("PC 1")
else:
scat[i] = axs.flatten()[i].scatter(
dihe_r[i][:, 0],
dihe_r[i][:, 1],
c=weights[i],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs.flatten()[i].set_title(
f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}"
)
axs.flatten()[i].set_xlabel("PC 1")
axs.flatten()[i].text(
-0.1,
1.15,
panel,
transform=axs.flatten()[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs.flatten()[0].set_ylabel("PC 2")
if multiple:
axs.flatten()[2].set_ylabel("PC 2")
colorbar = fig.colorbar(
scat[0], ax=axs, label="Energy [kcal/mol]", location="right"
)
else:
colorbar = fig.colorbar(
scat[0], ax=axs, label="Energy [kcal/mol]", location="right"
)
fig.savefig(
snakemake.output.report_pca_comparison, bbox_inches="tight", dpi=600
)
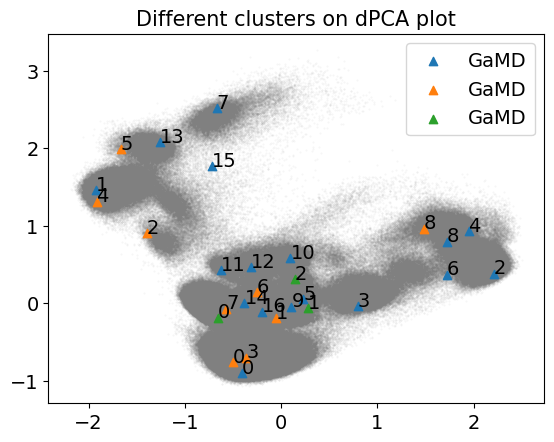
3.4.3. Compare MD clusters#
# Load cluster data
clusters = []
for i in range(no_md):
try:
clust_struct = md.load(snakemake.input[f"clusters_{i}"])
except:
clust_struct = None
clusters.append(clust_struct)
# Transform clusters in pca
clusters_dih = []
[
clusters_dih.append(src.dihedrals.getReducedDihedrals(clus))
for clus in clusters
]
cluster_pca = []
[cluster_pca.append(pca.transform(clus)) for clus in clusters_dih]
[None, None, None]
# Plot Clusters
fig, ax = plt.subplots()
for i in range(no_md):
ax.scatter(
dihe_r[i][:, 0],
dihe_r[i][:, 1],
marker=".",
s=0.5,
alpha=0.1,
c="grey",
rasterized=True,
)
# Plot clusters
for clus, method in zip(cluster_pca, list(methods.values())):
ax.scatter(clus[:, 0], clus[:, 1], marker="^", label=method)
for i, txt in enumerate(np.arange(len(clus[:, 0]))):
ax.annotate(
txt,
(
clus[i, 0],
clus[i, 1],
),
)
ax.legend()
ax.set_title("Different clusters on dPCA plot")
fig.savefig(snakemake.output.cluster_pca, dpi=600)
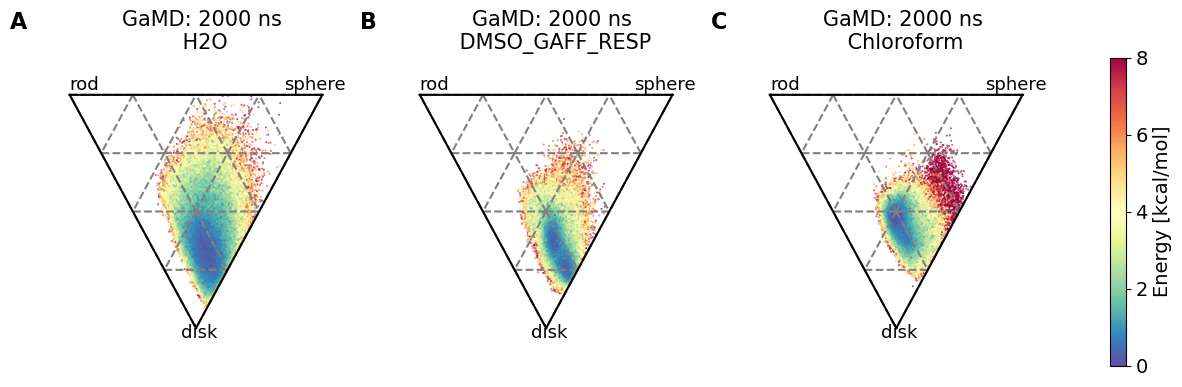
3.4.4. Shape comparison#
# Shape comparison plots - Load shapes
shape = {}
shape_weights = {}
for i in range(no_md):
shape[i] = src.utils.pickle_load(snakemake.input[f"shape_{i}"])
shape_weights[i] = src.utils.pickle_load(
snakemake.input[f"shape_weights_MC_{i}"]
)
fig, axs = plt.subplots(1, no_md, sharex="all", sharey="all")
fig.set_size_inches(16 / 3 * no_md, 4)
# create the grid
corners = np.array([[1, 1], [0.5, 0.5], [0, 1]])
triangle = tri.Triangulation(corners[:, 0], corners[:, 1])
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
outline = refiner.refine_triangulation(subdiv=0)
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
trimesh = refiner.refine_triangulation(subdiv=2)
scat = {}
for i, panel in enumerate(panel_labels):
scat[i] = axs[i].scatter(
shape[i][:, 0],
shape[i][:, 1],
c=shape_weights[i],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs[i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[i].set_xlabel(r"$I_{1}/I_{3}$")
axs[i].triplot(trimesh, "--", color="grey")
axs[i].triplot(outline, "k-")
axs[i].text(0, 1.01, "rod", fontsize=SMALL_SIZE)
axs[i].text(0.85, 1.01, "sphere", fontsize=SMALL_SIZE)
axs[i].text(0.44, 0.48, "disk", fontsize=SMALL_SIZE)
axs[i].scatter(0, 1.05, alpha=0, s=0.1)
axs[i].scatter(1.05, 1.05, alpha=0, s=0.5)
axs[i].scatter(0.5, 0.45, alpha=0, s=0.5)
axs[i].axis("off")
axs[i].text(
-0.1,
1.15,
panel,
transform=axs[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[0].set_ylabel("$I_{2}/I_{3}$")
colorbar = fig.colorbar(
scat[0], ax=axs, label="Energy [kcal/mol]", location="right", anchor=(0, 0)
)
# fig.tight_layout()
fig.savefig(
snakemake.output.shape_comparsion, dpi=600
) # bbox_inches='tight',
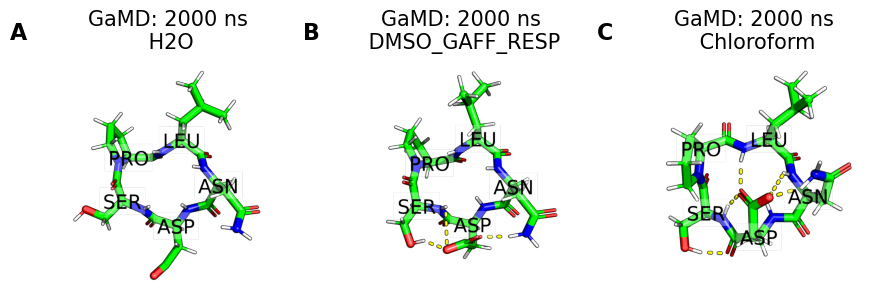
clusters = [
snakemake.input.clusters_0,
snakemake.input.clusters_1,
snakemake.input.clusters_2,
]
fig, axs = plt.subplots(1, no_md, figsize=(9, 4))
for i, panel in enumerate(panel_labels):
pymol_render = src.utils.pymol_image(
clusters[i], ref=snakemake.input.clusters_0, label=True
)
axs[i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[i].axis("off")
axs[i].text(
-0.1,
1.15,
panel,
transform=axs[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[i].imshow(pymol_render)
fig.tight_layout()
fig.savefig(snakemake.output.cluster_hbonds, dpi=600)
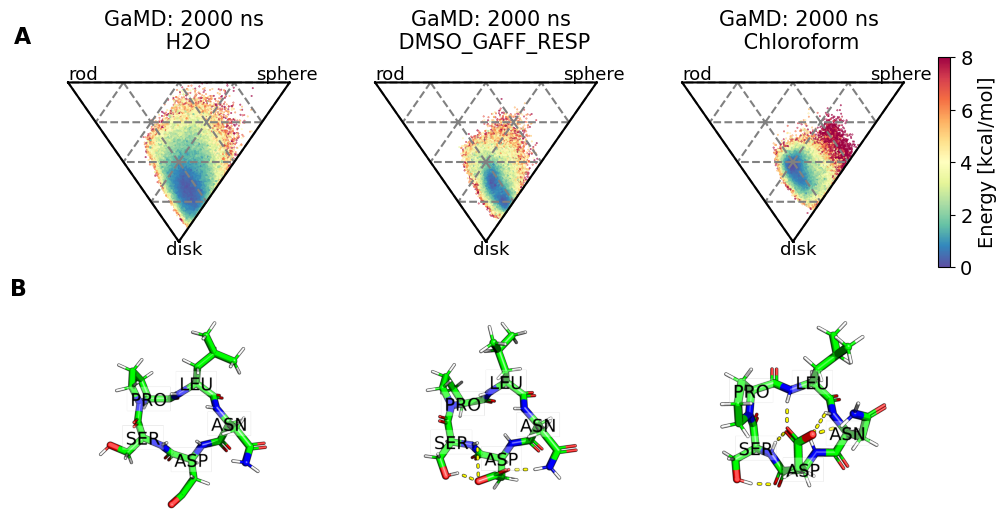
fig, axs = plt.subplots(2, no_md, sharex="row", sharey="row")
fig.set_size_inches(12 / 3 * no_md, 6)
# create the grid
corners = np.array([[1, 1], [0.5, 0.5], [0, 1]])
triangle = tri.Triangulation(corners[:, 0], corners[:, 1])
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
outline = refiner.refine_triangulation(subdiv=0)
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
trimesh = refiner.refine_triangulation(subdiv=2)
scat = {}
for i, panel in enumerate(panel_labels):
scat[i] = axs[0, i].scatter(
shape[i][:, 0],
shape[i][:, 1],
c=shape_weights[i],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs[0, i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[0, i].set_xlabel(r"$I_{1}/I_{3}$")
axs[0, i].triplot(trimesh, "--", color="grey")
axs[0, i].triplot(outline, "k-")
axs[0, i].text(0, 1.01, "rod", fontsize=SMALL_SIZE)
axs[0, i].text(0.85, 1.01, "sphere", fontsize=SMALL_SIZE)
axs[0, i].text(0.44, 0.46, "disk", fontsize=SMALL_SIZE)
axs[0, i].scatter(0, 1.05, alpha=0, s=0.1)
axs[0, i].scatter(1.05, 1.05, alpha=0, s=0.5)
axs[0, i].scatter(0.5, 0.45, alpha=0, s=0.5)
axs[0, i].axis("off")
axs[0, 0].text(
-0.1,
1.15,
"A",
transform=axs[0, 0].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[0, 0].set_ylabel("$I_{2}/I_{3}$")
clusters = [
snakemake.input.clusters_0,
snakemake.input.clusters_1,
snakemake.input.clusters_2,
]
for i, panel in enumerate(panel_labels):
pymol_render = src.utils.pymol_image(
clusters[i], ref=snakemake.input.clusters_0, label=True
)
# axs[i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[1, i].axis("off")
# axs[i].text(-0.1, 1.15, panel, transform=axs[i].transAxes,fontsize=16, fontweight='bold', va='top', ha='right')
axs[1, i].imshow(pymol_render)
axs[1, 0].text(
-0.25,
1.15,
"B",
fontsize=16,
transform=axs[1, 0].transAxes,
fontweight="bold",
va="top",
ha="right",
)
fig.subplots_adjust(right=0.85)
# get lower left (x0,y0) and upper right (x1,y1) corners
[[x10, y10], [x11, y11]] = axs[0, no_md - 1].get_position().get_points()
pad = 0.01
width = 0.01
cax = fig.add_axes([x11 + pad, y10, width, y11 - y10])
colorbar = fig.colorbar(scat[0], cax=cax, label="Energy [kcal/mol]")
fig.savefig(snakemake.output.cluster_hbonds, bbox_inches="tight", dpi=600) #
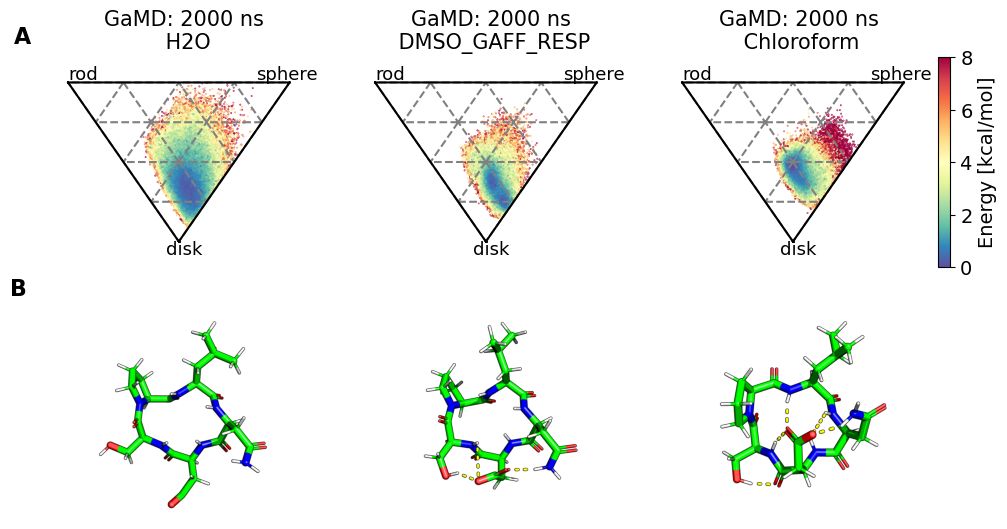
fig, axs = plt.subplots(2, no_md, sharex="row", sharey="row")
fig.set_size_inches(12 / 3 * no_md, 6)
# create the grid
corners = np.array([[1, 1], [0.5, 0.5], [0, 1]])
triangle = tri.Triangulation(corners[:, 0], corners[:, 1])
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
outline = refiner.refine_triangulation(subdiv=0)
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
trimesh = refiner.refine_triangulation(subdiv=2)
scat = {}
for i, panel in enumerate(panel_labels):
scat[i] = axs[0, i].scatter(
shape[i][:, 0],
shape[i][:, 1],
c=shape_weights[i],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=min_,
vmax=max_,
rasterized=True,
)
axs[0, i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[0, i].set_xlabel(r"$I_{1}/I_{3}$")
axs[0, i].triplot(trimesh, "--", color="grey")
axs[0, i].triplot(outline, "k-")
axs[0, i].text(0, 1.01, "rod", fontsize=SMALL_SIZE)
axs[0, i].text(0.85, 1.01, "sphere", fontsize=SMALL_SIZE)
axs[0, i].text(0.44, 0.46, "disk", fontsize=SMALL_SIZE)
axs[0, i].scatter(0, 1.05, alpha=0, s=0.1)
axs[0, i].scatter(1.05, 1.05, alpha=0, s=0.5)
axs[0, i].scatter(0.5, 0.45, alpha=0, s=0.5)
axs[0, i].axis("off")
axs[0, 0].text(
-0.1,
1.15,
"A",
transform=axs[0, 0].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[0, 0].set_ylabel("$I_{2}/I_{3}$")
clusters = [
snakemake.input.clusters_0,
snakemake.input.clusters_1,
snakemake.input.clusters_2,
]
for i, panel in enumerate(panel_labels):
pymol_render = src.utils.pymol_image(
clusters[i], ref=snakemake.input.clusters_0, label=False
)
# axs[i].set_title(f"{methods[i]}: {simtime[i]} ns\n {solvent[i]}")
axs[1, i].axis("off")
# axs[i].text(-0.1, 1.15, panel, transform=axs[i].transAxes,fontsize=16, fontweight='bold', va='top', ha='right')
axs[1, i].imshow(pymol_render)
axs[1, 0].text(
-0.25,
1.15,
"B",
fontsize=16,
transform=axs[1, 0].transAxes,
fontweight="bold",
va="top",
ha="right",
)
fig.subplots_adjust(right=0.85)
# get lower left (x0,y0) and upper right (x1,y1) corners
[[x10, y10], [x11, y11]] = axs[0, no_md - 1].get_position().get_points()
pad = 0.01
width = 0.01
cax = fig.add_axes([x11 + pad, y10, width, y11 - y10])
colorbar = fig.colorbar(scat[0], cax=cax, label="Energy [kcal/mol]")
fig.savefig(snakemake.output.cluster_hbonds_nolabel, bbox_inches="tight", dpi=600) #
3.4.5. Cheminformatics conformer generators (optional)#
snakemake.wildcards.confgens
'0_0'
# Load conformer generator structures
if snakemake.wildcards.confgens != "0_0":
confgen_number = int(len(snakemake.wildcards.confgens.split("_")) / 2)
confgens = snakemake.wildcards.confgens.split("_")
confgen_name = []
for idx, (j, k) in enumerate(zip(confgens[0::2], confgens[1::2])):
confgen_name.append(f"{j}:{k}")
chem_info_t = [
md.load(snakemake.input[f"cheminfoconfs{i}"])
for i in range(confgen_number)
]
print(chem_info_t)
chem_info_dihe_r = {}
chem_info_shapes = {}
for i, chem_t in enumerate(chem_info_t):
chem_info_dihedrals = src.dihedrals.getReducedDihedrals(chem_t)
chem_info_dihe_r[i] = pca.transform(chem_info_dihedrals)
# compute shape
inertia_tensor = md.compute_inertia_tensor(chem_t)
principal_moments = np.linalg.eigvalsh(inertia_tensor)
# Compute normalized principal moments of inertia
npr1 = principal_moments[:, 0] / principal_moments[:, 2]
npr2 = principal_moments[:, 1] / principal_moments[:, 2]
chem_info_shapes[i] = np.stack((npr1, npr2), axis=1)
else:
confgen_number = 1
if snakemake.wildcards.confgens != "0_0":
replacement_dict = {'omega:basic': "OMEGA Macrocycle", 'rdkit:ETKDGv3mmff': "RDKit ETKDG"}
for index, element in enumerate(confgen_name):
if element in replacement_dict:
confgen_name[index] = replacement_dict[element]
# Compare all cheminformatics conformer generators to md reference
all_panel_labels = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
]
n_row, n_col, figsize = src.utils.determine_no_plots(confgen_number)
fig, axs = plt.subplots(
n_row, n_col, sharex="all", sharey="all", figsize=figsize
)
if snakemake.wildcards.confgens != "0_0":
scat = {}
ref_label = f"{methods[0]}: {simtime[0]} ns\n {solvent[0]}"
for i in range(confgen_number):
scat[i] = axs.flatten()[i].scatter(
dihe_r[0][:, 0],
dihe_r[0][:, 1],
c=weights[0],
cmap="Spectral_r",
marker=".",
s=0.5,
vmin=0,
vmax=8,
rasterized=True,
label=ref_label,
)
scat[i] = axs.flatten()[i].scatter(
chem_info_dihe_r[i][:, 0],
chem_info_dihe_r[i][:, 1],
marker="s",
s=6,
color="black",
label=confgen_name[i],
) # , c=weights[i], marker='.', cmap='Spectral_r', s=0.5, vmin=min_, vmax=max_)
axs.flatten()[i].set_title(confgen_name[i])
axs[i].set_xlabel("PC 1")
axs.flatten()[i].legend(
bbox_to_anchor=(0, -0.4), loc="lower left", borderaxespad=0
)
axs.flatten()[i].text(
-0.1,
1.15,
all_panel_labels[i],
transform=axs[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[0].set_ylabel("PC 2")
fig.tight_layout()
fig.savefig(snakemake.output.all_cheminfo_comp_pca, dpi=600)
# Plot shapes
n_row, n_col, figsize = src.utils.determine_no_plots(confgen_number, 4, 3)
fig, axs = plt.subplots(
n_row, n_col, sharex="all", sharey="all", figsize=figsize
)
if snakemake.wildcards.confgens != "0_0":
# create the grid
corners = np.array([[1, 1], [0.5, 0.5], [0, 1]])
triangle = tri.Triangulation(corners[:, 0], corners[:, 1])
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
outline = refiner.refine_triangulation(subdiv=0)
# creating the outline
refiner = tri.UniformTriRefiner(triangle)
trimesh = refiner.refine_triangulation(subdiv=2)
scat = {}
for i in range(confgen_number):
scat[i] = axs.flatten()[i].scatter(
shape[0][:, 0],
shape[0][:, 1],
c=shape_weights[0],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=0,
vmax=8,
rasterized=True,
label=ref_label,
)
axs.flatten()[i].scatter(
chem_info_shapes[i][:, 0],
chem_info_shapes[i][:, 1],
marker=".",
c="black",
s=4,
label=confgen_name[i],
)
axs.flatten()[i].set_xlabel(r"$I_{1}/I_{3}$")
axs.flatten()[i].triplot(trimesh, "--", color="grey")
axs.flatten()[i].triplot(outline, "k-")
axs.flatten()[i].text(0, 1.01, "rod", fontsize=SMALL_SIZE)
axs.flatten()[i].text(0.85, 1.01, "sphere", fontsize=SMALL_SIZE)
axs.flatten()[i].text(0.44, 0.45, "disk", fontsize=SMALL_SIZE)
axs.flatten()[i].scatter(0, 1.05, alpha=0, s=0.1)
axs.flatten()[i].scatter(1.05, 1.05, alpha=0, s=0.5)
axs.flatten()[i].scatter(0.5, 0.45, alpha=0, s=0.5)
axs.flatten()[i].axis("off")
axs.flatten()[i].set_title(confgen_name[i])
axs.flatten()[i].legend(
bbox_to_anchor=(0, -0.4), loc="lower left", borderaxespad=0
)
axs.flatten()[i].text(
-0.1,
1.15,
all_panel_labels[i],
transform=axs[i].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs.flatten()[0].set_ylabel("$I_{2}/I_{3}$")
colorbar = fig.colorbar(
scat[0],
ax=axs,
label=" Energy [kcal/mol]",
location="left",
anchor=(8.5, 0),
)
# fig.tight_layout()
fig.savefig(snakemake.output.all_cheminfo_comp_shape, dpi=600)
fig, axs = plt.subplots(1, 2, figsize=(12, 4))
if snakemake.wildcards.confgens != "0_0":
colors = [
"black",
"red",
]
axs[0].scatter(
dihe_r[0][:, 0],
dihe_r[0][:, 1],
c=weights[0],
cmap="Spectral_r",
marker=".",
s=0.5,
vmin=0,
vmax=8,
rasterized=True,
)
for i in range(confgen_number):
axs[0].scatter(
chem_info_dihe_r[i][:, 0],
chem_info_dihe_r[i][:, 1],
c=colors[i],
marker="s",
s=6,
label=confgen_name[i], # capitalize()
) # markers
# bbox_to_anchor=(2.05,0), loc='upper left')
axs[0].set_xlabel("PC1")
axs[0].set_ylabel("PC2")
axs[0].text(
-0.1,
1.15,
all_panel_labels[0],
transform=axs[0].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
scat = axs[1].scatter(
shape[0][:, 0],
shape[0][:, 1],
c=shape_weights[0],
marker=".",
cmap="Spectral_r",
s=0.5,
vmin=0,
vmax=8,
rasterized=True,
)
for i in range(confgen_number):
axs[1].scatter(
chem_info_shapes[i][:, 0],
chem_info_shapes[i][:, 1],
c=colors[i],
marker=".",
s=4,
)
# axs[1].set_xlabel(r'$I_{1}/I_{3}$')
axs[1].triplot(trimesh, "--", color="grey")
axs[1].triplot(outline, "k-")
axs[1].text(0, 1.01, "rod", fontsize=SMALL_SIZE)
axs[1].text(0.85, 1.01, "sphere", fontsize=SMALL_SIZE)
axs[1].text(0.44, 0.45, "disk", fontsize=SMALL_SIZE)
axs[1].scatter(0, 1.05, alpha=0, s=0.1)
axs[1].scatter(1.05, 1.05, alpha=0, s=0.5)
axs[1].scatter(0.5, 0.45, alpha=0, s=0.5)
axs[1].text(
-0.1,
1.15,
all_panel_labels[1],
transform=axs[1].transAxes,
fontsize=16,
fontweight="bold",
va="top",
ha="right",
)
axs[1].axis("off")
# axs[1].set_title(snakemake.wildcards[f"confgen{fig_to_plot+1}"])
# axs[1].set_ylabel('$I_{2}/I_{3}$')
axs[0].legend(
mode="expand",
ncol=2,
loc="lower left",
bbox_to_anchor=(-0.1, -0.3, 1.5, 0.102),
borderaxespad=0,
)
colorbar = fig.colorbar(
scat, ax=axs, label="Energy [kcal/mol]", location="right"
)
fig.savefig(snakemake.output.single_comp_plot, bbox_inches="tight", dpi=600)